| Resources | Objectives | Procedures | Marks | |
| Kuby's Immunology |
Immunology Reference
Shelf |
Carney's Learning Techniques |
To Tortora's Microbiology |
Immunology Laboratory |
Using the Gen Bank Database
Updated 28/11/00
| Resources | Objectives | Procedures | Marks | |
| Kuby's Immunology |
Immunology Reference
Shelf |
Carney's Learning Techniques |
To Tortora's Microbiology |
Immunology Laboratory |
This Lab exercise contains a Description of the needed source materials, a list of objectives, a detailed description of each of the objectives and the marking scheme
OBJECTIVES:
Introduction
The mammalian immune system has innate and adaptive components. The function of the adaptive immune system relies on the ability of immune system to alter the amino acid sequences of specific proteins in order to better recognise and defend against specific pathogens. The sequences of these proteins is altered over several generations in the case of Major Histocompatibility Complex receptors , or over the course of a single infection in the case of Immunoglobulins and T-Cell receptors . The presence of many different forms of a specific protein within a population of animals is call polymorphism. Modern molecular biological and biochemical techniques allow researchers to study these amino acid differences by sequencing genes or proteins from different cells and individuals.and then comparing the sequences. In this laboratory you will obtain several sequences from one of the proteins mentioned above from an online database and analyse their amino acid sequences in order to determine the degree of polymorphism.
OBJECTIVE 1: Obtain amino acid Sequences from the GenBank Database
READ THIS ENTIRE OBJECTIVE BEFORE YOU START !!!
In this objective you will learn to access the National Center for Biotechnology Information website and use their "Entrez" search engine to find and retrieve protein sequences that will be used later in the exercise
To achieve this objective you will
The National Center for Biotechnology Information (NCBI) is an international repository of data on the sequence of genes and proteins. Researchers from around the world deposit their information here so that it can be used by the scientific community. The data are stored in several different databases; the "proteins" databases contain just the amino acid sequences while the "Nucleotides" databases contain both the nucleotide sequences and the deduced amino acid sequences.You can use either of these databases to retrieve the sequences that you will need for this exercise.
Data on millions of sequences are stored in separate files each with a unique identification code. You can use key-words to find specific sequences deposited in the databases.
The NCBI is constantly updated and modified as they find faster ways to retreive the information. Do not be surprised if the screen layout changes frequently as improvements ae brought online.
As you browse the NCBI website look for answers to the following questions
RETRIEVING AMINO ACID SEQUENCES
Before you start your search make certain that you have space on your computer to store the info that you retrieve. While you can store the info on a hard drive, a 3 1/2" floppy disk will be more than enough. As you will have to submit the data on a floppy disk anyway, why not save it directly to floppy disk?
Also, before you start your search make certain that you have opened a text file for future storage. Open a new text file in NotePad or Wordpad
Use the NCBI Entrez search engine to obtain 10 amino acid sequences of a specific protein from a single species and cut and paste these sequences in the opened text file as described below
You want to find specific sequences of proteins that are immunologically important eg, MHC, T cell Receptors, Kappa chain, Lambda chains or any of the heavy chains of immunoglobulins. Use only sequences from a single isotype, for example IgM or IgG, and from only the heavy or the light chain. The Entrez search engine lets you use key words to find these
Examples of Key word searches
After you have completed your search you will have a list of files with the appropriate sequences
SAVING THE SEQUENCE DATA
FORMATTNG THE TEXT FILE TO BE USED BY CLUSTAL X
The text file containing your sequences should be formatted to meet the following:
CLUSTAL FORMAT RULES
I usually call the file something that will indicate what I am aligning. The example below would be "stvi.txt" .
EXAMPLE OF A CLUSTAL FORMATTED TEXT FILE
>P1;Stvi-10-8
HSLKFYCTASSGVPNFPEFVAVGLVDDVEISHYDSNTKREEPKQDWMSRVTEDDPQFWQRETEISEGHQQTFKANIEVVRQRLNQT*
>P1;Auha-706
HSLKYFYTASSQVPNFPEFVTVGMVDDVQVDYYDSDTEKAEPKQDWIARNTDQQYWERNTDIYRGSQQSFKANIEIVKQRFNQT*
>P1;Auha-705
HSLKFFCTETPGVQSIPEFVAVAFVDEVQIGDFNNVRGAEPKKDWIKFFADHPEHLEWYSSISKQSHQVFKANIETFRQRLNQT*
>P1;Auha-517
HSLKYFFTGSSQVPNFPEFVVVGMVDDVQVVHYDSDTEKAGPKQDWFARNTDQQYWESQTGNLLGSQQTFKANIETAKQRFNQT*
>P1;Cyca-UAA
HTLQYFYTATSGIENFPEFMTAGVVDGQQIDYYDSIIRKAVQKAEWISGAVDPDYWNRNTQIYAGNEPSFKENINIVKSRFNQT*
>P1;Furu-UAA
HTLKYFYTASSGVPNFPEFVIVGMVDDVQMVRYDSNTRRMQPKQEWMKEFTADDPQYLDTQSQNAFGAQQIYKANIEIAKERFNQT*
>P1;Sasa-UAA
HALKYFYTASSEVPNFPEFVVVGVVDGVQMVHYDSNSQRAVPKQDWVNKAADPQYWERNTGIFKGSQQTFKANIDIAKQRFNQS*
>P1;Plam-sp005
HSLKNFHTASSQVTNIPEFVFVGFIDDVQTEYYDSNTKKSEPKQDWMLKATDAKYWERQTGILRAYQRVFKANMEIAKERFNQT*
>P1;Stvi-10-9
HSLKFYFTAFFGVPNFPEFVSVGLVDDIEISRYDSNTKREEPTQDWMSRVPQEDPPFWPTETEISDGDQQTFQAHIEVVKQRLNQT*
>P1;Stvi-10-10
HSLKIFCTACSGVPNFPEYVSVGLVDGVQMIRYDSNTKRQEPKQDWMSRVTEDDPQYWQRNTEIAEGNQQVYKAGIEILKQRLNQT*
Submit a copy of your file with your report.
OBJECTIVE 2 Install Clustal X on your Computer
DownLoad Clustal X
A copy of Clustal X is stored in
Immunology>>Resources
>>Software Downloads>> Clustal X under the file
name clustalx1_8msw.exe This is a compressed
document that has several files. This program will not work until
it has been downloaded and installed
To Download this file, goto the appropriate section on the website, Double click on the filename, tell the computer where you want to store the file on your hard drive. Remember where you have stored it!
Install Clustal X
To install Clustal X, find the file clustalx1_8msw.exe
that is now stored on your computer. Double click on the
file name and follow the install instructions You will have to
tell the computer where you want the program ClustalX installed.
You could create a subdirectory called Clustal in the Program
Files directoryand install CLUSTALX in the subdirectory.An
executable file (clustalx.exe) which will run
under MS Windows (32 bit). The directory containing the
executable (plus the files named *.par,and clustalx.hlp) should
be added to your path defined in the autoexec.batfile.
Browse through the menu options and the help files of ClustalX to learn some of its features
 |
The BioBox website compares both the Clustal W and the Clustal X programs. The Clustal W is a DOS version. The Clustal X is a windows version |
| http://www.csc.fi/molbio/progs/clustalw/ |
OBJECTIVE 3 Align the sequences using Clustal X
download the file called Clustal_Example.txt an use it as a test example .
Start the program Clustal X (clustalx.exe).
Select Multiple Alignment Mode
Select file
select load sequences
Find and select the file Clustal_Example.txt If ClustalX recognises you file, it will list the sequences and the number of amino acids in each one. (If it doesn’t do this, check that your file is saved as text only).
The path and the filename should appear at the bottom of the screen
A list of the alignment sequences will appear in the left hand frame
The sequences will appear as multi coloured letter codes on the right hand frame
Select the aligment option from the menu
Selct the Do Complete Alignment option
You will be asked for names for the alignment and dendrogram files. Just use the default names, which will be the same as your text file, with either an .aln (eg stvi.aln) or .dnd (eg stvi.dnd) extension. The alignment will appear on the screen.
Close the clustal window, then using a wordprocessor such as Microsoft Wordpad, Microsoft Word or Wordperfect open up the .aln file (eg stvi.aln). Your alignment should be there and it should look like the example below (note the example below is comparing MHC class I sequences from several different fish species - your sequences should be much more similar!). Deletions within the sequence will be indicated by dashes. The bottom line of the alignment will contain several symbols. Stars (*) indicate amino acids which are identical, colons (:) indicate amino acid changes which are conservative (hydrophobic for hydrophobic or acidic for acidic), while periods (.) indicate changes which may be conservative. If the rows do not line up, change the font to Courier, 10 point.
CLUSTAL X (1.8) multiple sequence alignment
Stvi-10-8
HSLKFYCTASSGVPNFPEFVAVGLVDDVEISHYDSNTKREEPKQDWMSRVTEDDPQFWQR
Stvi-10-9
HSLKFYFTAFFGVPNFPEFVSVGLVDDIEISRYDSNTKREEPTQDWMSRVPQEDPPFWPT
Stvi-10-10
HSLKIFCTACSGVPNFPEYVSVGLVDGVQMIRYDSNTKRQEPKQDWMSRVTEDDPQYWQR
Cyca-UAA
HTLQYFYTATSGIENFPEFMTAGVVDGQQIDYYDSIIRKAVQKAEWISG--AVDPDYWNR
Sasa-UAA
HALKYFYTASSEVPNFPEFVVVGVVDGVQMVHYDSNSQRAVPKQDWVNK--AADPQYWER
Auha-706
HSLKYFYTASSQVPNFPEFVTVGMVDDVQVDYYDSDTEKAEPKQDWIAR--NTDQQYWER
Auha-517
HSLKYFFTGSSQVPNFPEFVVVGMVDDVQVVHYDSDTEKAGPKQDWFAR--NTDQQYWES
Plam-sp005
HSLKNFHTASSQVTNIPEFVFVGFIDDVQTEYYDSNTKKSEPKQDWMLK--ATDAKYWER
Furu-UAA
HTLKYFYTASSGVPNFPEFVIVGMVDDVQMVRYDSNTRRMQPKQEWMKEFTADDPQYLDT
Auha-705
HSLKFFCTETPGVQSIPEFVAVAFVDEVQIGDFN-NVRGAEPKKDWIKFF-ADHPEHLEW
*:*: : * : .:**:: ...:* : :: . . :*. . .
Stvi-10-8
ETEISEGHQQTFKANIEVVRQRLNQT
Stvi-10-9 ETEISDGDQQTFQAHIEVVKQRLNQT
Stvi-10-10 NTEIAEGNQQVYKAGIEILKQRLNQT
Cyca-UAA NTQIYAGNEPSFKENINIVKSRFNQT
Sasa-UAA NTGIFKGSQQTFKANIDIAKQRFNQS
Auha-706 NTDIYRGSQQSFKANIEIVKQRFNQT
Auha-517 QTGNLLGSQQTFKANIETAKQRFNQT
Plam-sp005 QTGILRAYQRVFKANMEIAKERFNQT
Furu-UAA QSQNAFGAQQIYKANIEIAKERFNQT
Auha-705 YSSISKQSHQVFKANIETFRQRLNQT
: . :: :: :.*:**:
OBJECTIVE 4: Analyse the polymorphism in the aligned sequences
Researchers who study polymorphic molecules use a special method to present and interpret their results, called a Wu-Kabat variability plot. This is a graph which indicates the position along the amino acid chain on the X axis and the variability index along the Y axis. The variability index is calculated as follows:
Number of different amino acids at a given position along the amino acid chain
Frequency of the most common amino acid present at a given position along the amino acid chain
For example in the alignment above position 1 is histidine (H) in all ten sequences so the variability index is calculated as follows:
Number of different amino acids = 1
Frequency of the most common =10 out of 10 = 10/10 = 1
Variability index = 1/1 = 1
In the alignment above position 2 is variable and the variability index is calculated as follows:
Number of different amino acids (S, T, A) = 3
Frequency of the most common (S)= 7 out of 10 = 7/10 = 0.7
Variability index = 3/0.7 = 4.29
Position 21, which follows the sequence PEFV, is variable and the variability index is calculated as follows:
Number of different amino acids (A,S,T,V,F,I)= 6
Frequency of the most common (A or S or T or V) =2 out of 10 = 2/10 = 0.2
Variability index = 6/0.2 = 30
In a Wu-Kabat plot these values are plotted against their position in the polypeptide chain. An example of such a plot is given below:
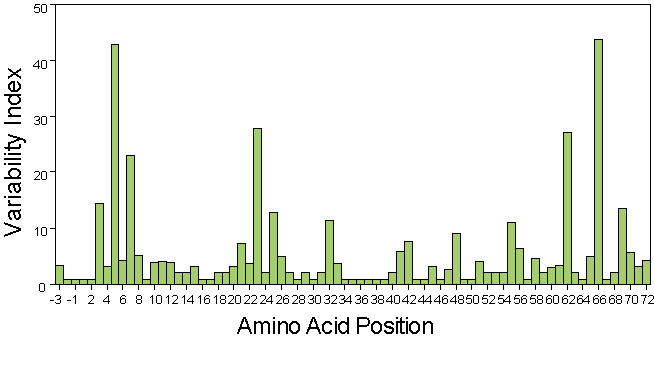
OBJECTIVE 5: Submit a Report
Construct a Wu-Kabat plot for the alignment you have produced. Submit a report indicating the type of sequence you analysed (MHC, immunoglobulin, T- Cell receptor) and the names of the sequences you used. Include the your .pir file, your .aln file and your Wu-Kabat plot. You can hand in all of these items in electronic format on a 3.5 inch computer disk if you wish.
Appendix: The one letter amino acid code
CLUSTAL REFERENCES
Details of algorithms, implementation and useful tips on usage of
Clustal
programs can be found in the following publications:
Jeanmougin,F., Thompson,J.D., Gouy,M., Higgins,D.G. and
Gibson,T.J. (1998)
Multiple sequence alignment with Clustal X. Trends Biochem Sci,
23, 403-5.
Thompson,J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and
Higgins,D.G. (1997)
The ClustalX windows interface: flexible strategies for multiple
sequence
alignment aided by quality analysis tools. Nucleic Acids
Research, 24:4876-4882.
Higgins, D. G., Thompson, J. D. and Gibson, T. J. (1996) Using
CLUSTAL for
multiple sequence alignments. Methods Enzymol., 266, 383-402.
Thompson, J.D., Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W:
improving the
sensitivity of progressive multiple sequence alignment through
sequence
weighting, positions-specific gap penalties and weight matrix
choice. Nucleic
Acids Research, 22:4673-4680.
Higgins,D.G., Bleasby,A.J. and Fuchs,R. (1992) CLUSTAL V:
improved software for
multiple sequence alignment. CABIOS 8,189-191.
Higgins,D.G. and Sharp,P.M. (1989) Fast and sensitive multiple
sequence
alignments on a microcomputer. CABIOS 5,151-153.